19. The (Diagrammatic) Narratives of Genetic Revolutions
© 2024 Marianne Sommer, CC BY-NC-ND 4.0 https://doi.org/10.11647/OBP.0396.23
Historians of science Elsbeth Bösl and Elizabeth Jones have written about the development of aDNA research in cycles of proclaimed revolutions. The first cycle commenced with the search for the oldest DNA using polymerase chain reaction technology in the 1990s; this was followed by the second hype cycle in the field, with the advent of next-generation sequencing in this millennium (Jones and Bösl 2021; see also Bösl 2017). Here, I am particularly interested in how the continuities and breaks with classical population genetics were narrated. In a video on YouTube (2018a) as well as in his popular book of the same title, Who We Are and How We Got Here (2019), the influential American aDNA researcher David Reich (prominent both within science and in a broader public context) explained that he had been brought up intellectually in the tradition of Cavalli-Sforza:
This book is inspired by a visionary, Luca Cavalli-Sforza, the founder of genetic studies of our past. I was trained by one of his students, and so it is that I am part of his school, inspired by his vision of the genome as a prism for understanding the history of our species. (Reich 2019, xvii)
As we have seen, Cavalli-Sforza had been among those who brought about ‘the first revolution’ in human population-genetic studies when reconstructing modern human population history based on allele frequencies and later DNA sequences. A broad consensus was reached that this history could be modelled as a tree with its young root in Africa, from where humans successively populated the globe, splitting into independent lines.
However, according to Reich, “Cavalli-Sforza’s maps” (2019, xix) were wrong. The second, “ancient DNA revolution” (ibid.) showed that the present-day genetic structure of human populations was not sufficient to reconstruct ancient events because, contrary to Cavalli-Sforza’s expectations, people had mixed and blurred the genetic patterns of the past, and there had been major migrations to the effect that people occupying a particular geographic region today might not be entirely representative of, or descended from, those who lived there in the past. Cavalli-Sforza seems not to have been right in his assumption that the past was a much simpler place than the present. His reliance on what he thought to be isolated populations that provided a direct link into history was misguided (also Reich 2019, 219, 259).
So even though Reich honored the legacy of the early pioneers as transmitted in The History and Geography of Human Genes (Cavalli-Sforza, Menozzi, and Piazza 1994), he moved “[t]oward a new history and geography of human genes informed by ancient DNA” (Pickrell and Reich 2014, title). Reich referred to the founder of the new field as Svante Pääbo, who with his colleagues had developed genome-wide aDNA analyses that eventually gave them access to Neanderthal and Denisovan genomes. Reich was invited to collaborate with Pääbo in 2007, who was at the Max Planck Institute for Evolutionary Anthropology in Leipzig (Germany). This was due to the fact that Reich and the mathematician and computational geneticist, Nick Patterson, Reich’s close collaborator since the early 2000s, had made advances in the study of population admixtures. When research results began to point towards interbreeding of Neanderthals and modern humans, they were skeptical. Pääbo had received postdoctoral training in the very genetics laboratory out of which had come the decisive results to initiate a consensus around the out-of-Africa model that excluded interbreeding with archaic humans. Reich himself was biased against the notion that modern humans interbred with Neanderthals due to his having been emersed in ‘the Cavalli-Sforza paradigm’ that, too, was built on the out-of-Africa model (Reich 2019, 36). Yet, from their aDNA research, a picture emerged in which modern-human–Neanderthal hybrid populations had once occupied Europe and lived across Eurasia, many of which had died out, but some left behind genetic traces in present-day humans.
After seven years of spending time in Pääbo’s laboratory, Pääbo helped Reich install his own, the first to focus on the study of whole ancient human genomes in the US. Within a few years, more than half of the published genome-wide aDNA came from Reich’s laboratory at Harvard University. Reich was part of the endeavor that led to the overthrow of
the ‘serial founder effect’ models [7,8], which proposed that populations have remained in the locations they first colonized after the out-of-Africa expansion, exchanging migrants only at a low rate with their immediate neighbors until the long-range migrations of the past 500 years [9–12]. (Pickrell and Reich 2014, 377)
Reich was among those who showed that “[i]nstead, the past 50’000 years of human history have witnessed major upheavals, such that much of the geographic information about the first human migrations has been overwritten by subsequent population movements” (378). Most significantly, it became clear that “new types of models – with admixture at their center – are necessary for describing key aspects of human history […]” (ibid.).
In accordance with this move to new models, Reich verbally deconstructed the tree model: “The avalanche of new data that has become available in the wake of the genome revolution has shown just how wrong the tree metaphor is for summarizing the relationship among modern human populations” (2019, 77). Indeed, Reich went on to echo statements made by much earlier opponents of the tree model for human evolution, whom we met in Part III, like Julian Huxley in the 1930s. Instead referring to his contemporary Alan R. Templeton whom I discuss at the end of this chapter, Reich wrote that
[…] while a tree is a good analogy for the relationships among species – because species rarely interbreed and so like real tree limbs are not expected to grow back together after they branch – it is a dangerous analogy for human populations […] Instead of a tree, a better metaphor may be a trellis, branching and remixing far back into the past. (Reich 2019, 81)
Indeed, Reich stated that “[t]here was never a single trunk population in the human past. It has been mixtures all the way down” (2019, 82). At the same time, he referred to the f-statistics and D-statistics they used in testing for mixing that “evaluate whether a tree model is an accurate summary of real population relationships” (78). In fact, in a paper the first signatory of which was Patterson, they wrote that
“[t]hese methods are inspired by the ideas by Cavalli-Sforza and Edwards (1967) [a paper I discussed in Chapter 16], who fit phylogenetic trees of population relationships to the Fst values measuring allele frequency differentiation between pairs of populations” (Patterson et al. 2012, 1065–1066). Patterson, who has co-led the David Reich Lab for some twenty years and has been the driving force behind the mathematical and computational developments (e.g., the software in the package ADMIXTOOLS, Patterson et al. 2012), described Cavalli-Sforza as “a hero for David and me” (personal interview with Nick Patterson, 15 August 2023).
So there is testimony to the handing down of ideas, models, and diagrams of human history and kinship from geneticist to geneticist and laboratory to laboratory and of a few dominant and more or less “vertically integrated” laboratories with the necessary financial, technical, and human resources to carry out successful aDNA research (Pickrell and Reich 2014, 385; personal interview with Nick Patterson, 15 August 2023). This might be part of the reason why despite the rhetoric against the tree and in favor of the trellis, what was built were trees with a few connecting branches or arrows. Rather than models that express that “changes in populations over time are typically gradual – owing to consistent, low-level gene flow between neighboring populations”, we find visualizations of admixture and introgression as “punctate, with migration events rapidly altering the genetic composition of a region” (Pickrell and Reich 2014, 382–83).
This contradiction is paralleled by another regarding the nature of these populations. Reich declared that aDNA research had proven wrong the assumption of many people “that humans can be grouped biologically into ‘primeval’ groups, corresponding to our notion of ‘races’, whose origins are populations that separated tens of thousands of years ago” (Reich 2019, xxviii). Rather, aDNA research revealed that human diversity had changed radically in the course of evolution, so that today’s populations are complex admixtures of populations from the past which themselves were admixed. However, even though Reich wrote of “our interconnected human family” (22), the diagrams remained tree-like. Furthermore, despite the ‘admixture-instead-of-races’ narrative, he regarded it as “undeniable that there are nontrivial average genetic differences across populations in multiple traits” (253) and that “[t]he average time separation between pairs of human populations since they diverged from common ancestral populations […] is far from negligible on the time scale of human evolution” (258, also 265). In fact, Reich has been at the center of a controversy about the issue of race in science that has, among other things, led to a critical statement signed by sixty-seven researchers (Opinion, BuzzFeed 2018). The critique has not only been triggered by Reich’s Who We Are and How We Got Here (2019), but mainly by a provocative opinion Reich penned in The New York Times, urging people to take seriously the differences between human populations that scientific research had and would make known (Reich 2018b; particularly far-reaching claims regarding differences between human ‘races’ on the basis of population genetics have been made by the journalist Nicholas Wade [2014]).
This leads to another aspect of the stories population geneticists tell. It has to do with the controversy between out-of-Africa proponents and multiregionalists that predates the molecular approach to human evolutionary history (see Part III). While, as we have seen, the first scenario is organized around a relatively late radiation of modern humans from Africa across the globe, the rival multiregional model stresses the significance of local continuity in human evolutionary history. In the latter view, local Homo erectus populations gave rise to the modern human geographical varieties. Rather than assuming a relatively recent last common ancestor, and thus a human ‘racial divergence’ as recent as to coincide with Homo sapiens, the process is regarded as reaching further back, to when the supposedly first migrations out of Africa were undertaken by Homo erectus. In this scenario, Homo sapiens evolved locally from archaic populations that stood in genetic exchange with each other. Hence, the multiregional hypothesis is associated with less taxonomic diversity in the hominid record, and with a more linear phylogeny, including local Neanderthal populations as ancestral to modern humans.
According to narratives of some of today’s geneticists, it was human population genetics that refuted the multiregional model in the 1980s when it lent support to the out-of-Africa or mitochondrial Eve model, but then came the second, aDNA-driven revolution, and the pure out-of-Africa model was overthrown (e.g., López, Van Dorp, and Hellenthal 2015, 57–59; Reich 2019, 4–5; Gokcumen 2020, 61–62; Gopalan et al. 2021, 200). But what exactly do the findings coming out of the aDNA laboratories mean for the controversy? The answer to this question varies between researchers, and it is not always given in its full complexity. For even without the inclusion of aDNA, there was already a wealth of scenarios that differed in the location of modern human origins in Africa, the number of migrations and dispersals out of Africa, the route(s) taken, and in whether there were migrations back into Africa, as well as in the timing of events and the assumed amount of gene flow (see, e.g., López, Van Dorp, and Hellenthal 2015).
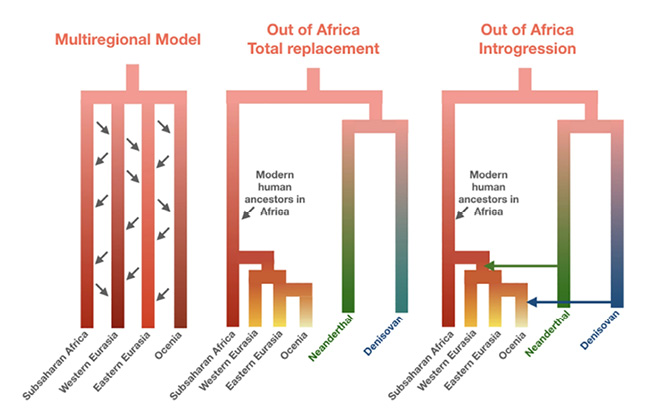
Fig. IV.19 “the three broad models of recent human evolution”. Omer Gokcumen, “Archaic Hominin Introgression into Modern Human Genomes” (Yearbook of Physical Anthropology 171.S70 [2020]: 60–73), Fig. 1, p. 63. © 2019 American Association of Physical Anthropologists, all rights reserved (reproduced with permission by John Wiley and Sons and Copyright Clearance Center).
Furthermore, a model similar to those that emerged from aDNA studies was also developed from morphological evidence for introgression of archaic humans into modern humans in Eurasia, and it was referred to by its creator, the paleoanthropologist Fred H. Smith, as the assimilation model. Interestingly, Smith and colleagues reproduced the figure from Prüfer et al. (2014, Fig. 8, 48) (see Figure IV.14 above) as an affirmation of their assimilation model, confirming that the tree has ‘simply’ acquired a few thin connecting arrows (Smith et al. 2017, 132–33; on this model, see also Ackermann et al. 2019). Nonetheless, in continuation of what I have found in Chapter 13 of Part III, a simplified historical succession of dominant models may be recounted in a series of diagrams, as in the case of Figure IV.19: the out-of-Africa model won over the multiregional model in the 1980s with the comparison of mtDNA from living individuals of different human populations, only to be replaced by a more complicated, ‘reticulate model’ with the aDNA studies that suggested introgression from archaic forms.
Thus, in the telling of the history of human population genetics at the time of aDNA research, diagrammatic means similar to those we have observed in Part III are still used, only that the aDNA model now triumphs at the end: to the left is the multiregional model with a linear diagram standing for anagenesis, in the middle the out-of-Africa model with a tree-shaped phylogeny standing for cladogenesis, and to the right is the reticulate model with a tree that shows connecting darts. Accordingly, the reticulate model is called “Out of Africa Introgression” model in Figure IV.19. It has also been referred to as the leaky replacement model, meaning an out-of-Africa model in which complete replacement of archaic forms by modern humans outside Africa has given way to the assumption of low degrees of introgression from archaic into modern populations (e.g., Nielsen et al. 2017; as we have seen, this has more recently been supplemented with the replacement of the mtDNA and Y chromosome in Neanderthals by modern humans [Liu et al. 2021]). In sharp contrast to a tradition I have documented in Part III, which tended to equate Weidenreich’s phylogenetic network with a candelabra model of parallel hominin and ‘racial’ evolution, Omer Gokcumen (2020) of the Evolutionary and Anthropological Genomics Laboratory at the University at Buffalo does justice to the multiregional model that can be seen as originating in Weidenreich’s diagram by including arrows to symbolize gene flow throughout hominin evolution.
Nonetheless, some researchers think the aDNA revolution was more significant. Reich actually wrote of a synthesis between the out-of-Africa and multiregional models, and he conceded that the species status of the Neanderthals, as well as the Denisovans, had become uncertain through the establishment of local interbreeding with modern humans (2019, 49–50, 56). Besides Africa, eastern Eurasia was confirmed as an important region of human evolution, and the picture that emerged of hominin existence some 70,000 years ago increased in complexity, with highly diversified groups like the Neanderthals and Denisovans, and the small Flores forms from today’s Indonesia, living at the same time and to some degree in contact with modern humans (62–64). Also ‘the beginning of our story in Africa’ has come to be presented differently in some papers, to the degree that an ‘African multiregionalism’ is proposed, with a gradual and mosaic development of ‘modern human traits’ among differentiated Pleistocene groups that were distributed across the continent and interconnected by gene flow in various degrees. In fact, in this view, and as I will explore further in Chapter 20, the biological and paleoanthropological species concept as well as the notions of ‘archaic’ and ‘modern’ need to be reconsidered. Simple tree-like demographic models for Africa, even if they include gene flow between branches, appear untenable to some researchers (Scerri et al. 2018; Galway-Witham, Cole, and Stringer 2019; Vincente and Schlebusch 2020). Instead, reminiscent of Julian Huxley’s verbal images encountered in Part III, the diagrammatic metaphor of a braided river has been suggested for modern human origins in Africa (Yong 2018). Thus,
[c]onsidering the increasing number of ancient individuals identified with recent archaic ancestries, past hominins may have mixed frequently, opening the question of whether archaic and modern human should be regarded as distinct lineages or rather points taken from a continuous spectrum of genetic diversity that was genetically connected throughout the past ~500 kyr similar to that of present-day human populations (20, 22, 23). (Liu et al. 2021, 1479)
At the same time, it seems that the picture aDNA studies have generally painted is one of complex population (pre)histories, with migration, mixture, replacement, and extinction repeatedly changing the genomic structure, including in Africa, rather than being one of local continuity, as in the multiregional model (Klein 2019; for an inclusion of fossil evidence, see also Stringer 2014). That the so-called revolution was not really overturning the treeness of human evolution and kinship may be illustrated by a summary diagram from the review paper that provided the above quote. It includes “[m]ultiple pulses of introgression” as well as the replacement of Neanderthal mtDNA and Y (Liu et al. 2021, Fig. 2, 1482, quote from caption) (see Figure IV.20). It is an orderly picture that, rather than conveying the explosive potential of subverting categories such as human ‘races’ or even hominin species, suggests neat separations.
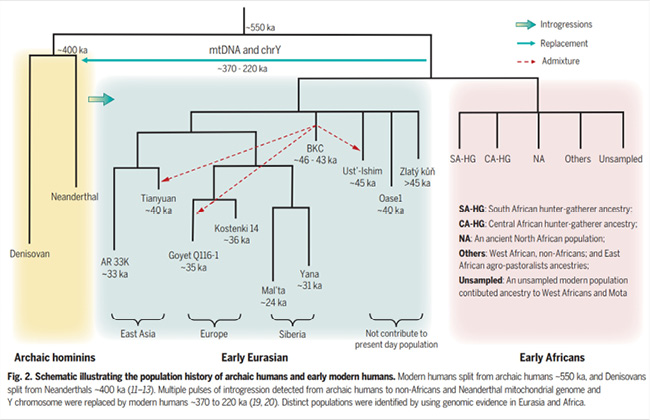
Fig. IV.20 “Schematic illustrating the population history of archaic humans and early modern humans”. From Yichen Liu, Xiaowei Mao, Johannes Krause, et al., “Insights into Human History from the First Decade of Ancient Human Genomics” (Science 373.6562 [2021]: 1479–84), Fig. 2, p. 1482. Reprinted with permission from AAAS © The American Association for the Advancement of Science, all rights reserved (“Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher”).
Both narratives, the one of human genetic history and the one of the history of human population genetics, have been told in different ways by those sympathetic towards the multiregional model. Nonetheless, they may use the same diagrammatic language as found in Figure IV.19 (e.g., Templeton 2018a, Fig. 6.2, 115, modified from Fagundes et al. 2007, Fig. 1, 17615, which is actually a paper in support of the out-of-Africa model). Researchers like the American geneticist and statistician Templeton agree that the pure out-of-Africa model that assumes total replacement of archaic by modern humans in Eurasia has been proven wrong. But in 2013, he considered something like a downsized multiregional model the best fit to existing data. Templeton criticized not only the pure out-of-Africa model with its denial of any interbreeding between modern and archaic humans but also the admixture-models that only allow for minor interbreeding (events). To him, human evolution has been dominated by gene flow and admixture, which has upheld humanity as a single evolutionary lineage. He did not accept the scenario according to which populations developed in relative isolation from one another (and only mixed at a later stage). In his view, the family tree – with or without connecting arrows – is no adequate model of human evolution for any period. One might rather have to think of a trellis (Templeton 2013, 267–70 on the trellis, Fig. 3, 268, for an image; see also Finlayson 2013).
Multiregionalists see genetic as well as archeological and paleoanthropological data as compatible with a model that combines significant migration and distribution events with regional lines of descent and gene flow between regions (mediated by isolation by distance). This is regarded as having maintained human variation within one species, including the archaic human forms like Homo erectus, Neanderthals, or Denisovans (e.g., Wolpoff 2020). Although such models are reminiscent of Weidenreich’s network of humanity from the 1940s (see Part III), the multiregional models have rather become a compromise between aspects of the tree (on a map) and the uniform (geographic) network. The respective diagrams contain visual elements of both. An early diagram by Templeton (2005, Fig. 9, 50) told of three migrations out of Africa (arrows), beginning with Homo erectus. The image also showed regional evolutionary developments (vertical lines) that were interconnected through gene flow (diagonal lines) (see Sommer 2015a, 135, 137). In the newer version of this diagram represented in Figure IV.21, Templeton’s trellis of human evolution appears more tree-shaped in the lowest part. The thin diagonal lines indicating gene flow between regions no longer go all the way down to Homo erectus, and with regard to the most recent expansion out of Africa, the model is described as mostly out-of-Africa with limited admixture (Templeton 2018b, Ch. 7, containing Fig. 7.4, 207; see also Templeton 2018a, Fig. 6.3, 120; Wadell 2018, in the same volume, arrived at the conclusion that the original out-of-Africa model has been mainly supported by new findings).
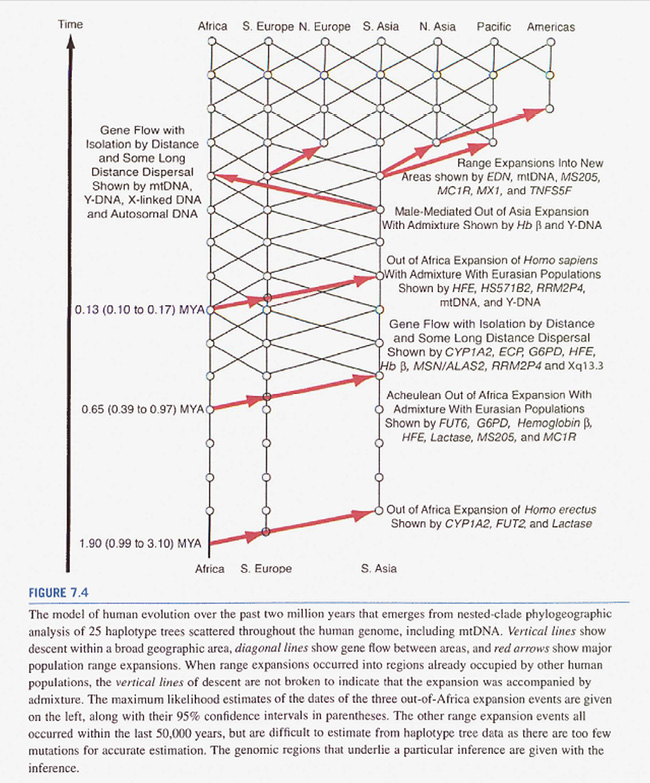
Fig. IV.21 “model of human evolution”. All rights reserved; used with permission of Elsevier Inc., from Alan R. Templeton, Human Population Genetics and Genomics (San Diego: Elsevier Science & Technology, 2018), Fig. 7.4, p. 207; permission conveyed through Copyright Clearance Center, Inc.
The diagrammatic changes from 2005 to Figure IV.21 in 2018 actually lead back to an important aspect of statistics that has come up several times before: hypothesis testing. Templeton had developed a method for testing the null hypothesis of no gene flow between two geographic regions from some time in the past up to the present. This test significantly indicated gene flow in the early Pleistocene, but it could not be ruled out that the result was affected by more recent genetic exchange. In 2009, he developed a more refined test to check the null hypothesis of no gene flow in a specific time interval in the past. This more refined test showed highly significant gene flow among human populations from the mid-Pleistocene onwards, but could not reject the null hypothesis of no gene flow for the early Pleistocene. Since Templeton’s diagrams were based upon statistically significant rejections of null hypotheses, he removed the trellis structure in the early Pleistocene. Though there was evidence of gene flow in that period, it was insufficient to reject the null hypothesis of no gene flow (personal communication, 8 January 2024).
At the same time, the mixture of modern and archaic humans in Eurasia had gained strong support from aDNA studies. Templeton saw in the results from aDNA studies further refutation of the population-tree model of human evolution. They confirmed network or trellis models that foreground gene flow and admixture since at least the mid-Pleistocene. The trees and their concomitant assumptions were “artifacts of using computer programs that force a treelike structure upon the data even if the data are not treelike” (2018a, 227). Similarly, Templeton regarded the genetic differences between Neanderthals, Denisovans, and modern humans to be insufficient to even regard them as subspecies (226). Diagrams such as given in Figure IV.21 therefore clearly testify to alternative ways of envisioning hominin diversity and kinship, and there is an increasing number of contenders in the race to (diagrammatically) re-define humans and human relatedness. To these revolutionary issues I now turn in my final chapter of this last part of the book.
